Siansivirga zeaxanthinifaciens CC-SAMT-1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Siansivirga; Siansivirga zeaxanthinifaciens
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
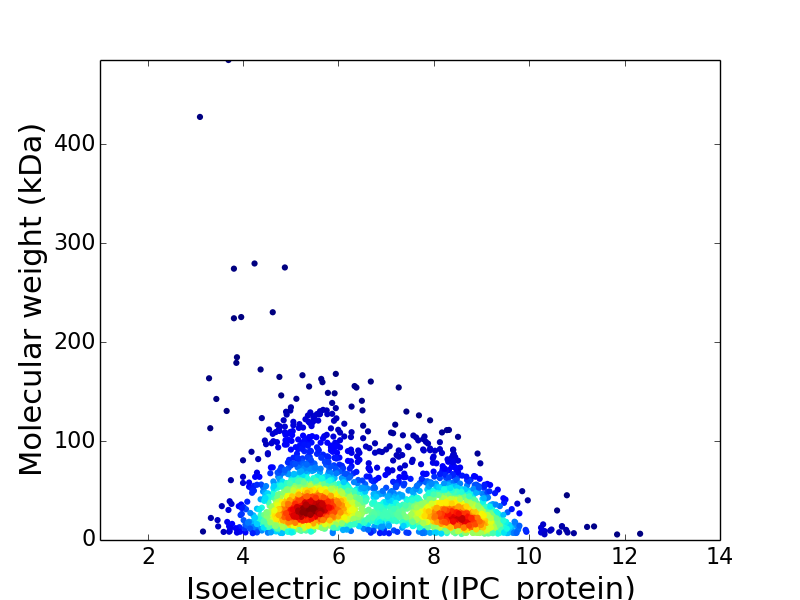
Virtual 2D-PAGE plot for 2,744 proteins (isoelectric point calculated using IPC_method)
Get csv file with sequences according given criteria:
* You can choose from 18 different methods for calculating isoelectric point
Protein with the lowest isoelectric point:
>tr|A0A0C5WI01|A0A0C5WI01_9FLAO Uncharacterized protein
MKKNFILIFLFLTSLIGNAQQQSEDCIVPASLTLTHFNTAKYVPGGHLAVFFEPEGVFELDNEFVLELSDASGSFSTATTLSMKSEFFIPVLNGIIPLDIAPGSNYKLRIRTTAPFASVETDAFEIISQTTPDVVPAEITFINNAFTDLDNFIKCVDLAHDNYFFGHKDKGLTEVTPSDGGGIKLGLLGG
NSSNTVVRMLINGLWQELSITTIGSQFTIPSGLPVGYYLMELDKTVNPNTPNEYHNISGFIFHFNTNITGIANTSSETICVDQNAGFEVPIASIQYNYPGSMYSIHYGDSDETDTSTFDFYTHARLVNCNTLNHVYNATTCSSIFQDENPGNDNFYFKLDFNLYSEGLFNSDSDQFECETYTKNGGGTTK
WINASLKPSADLIAPDIICEGLAIVATDNSTSGLYGFGPECSSDYNTYWEVAAPGDDWVSVDPNNSIFTGWIDDVARTLTIPGSITSNNPGFWGIRLIISNPLGCVQSDTDEQTVIVEPIPAPSFDYTYQDNLCAPVTIEFTNTSNTEDINPPSFGNPTYTWSVVPDLTTPASLNGFTLLDENPNDNIDA
ANQTDIDIRFTQPGTYIVTLQLENECGTIFFPQNITIIGDPTVSFNPNSLEVCQEAPANYTLDFSDSTIQPLYSDTPYTPTTFLWEVFEIDGVTPASNYTFVNSTDAGTNYPQINFTEFGEYLIRITVSGNCEGNGSDDFLFNLKQTPIITNTNLTQEICSGDDTDEILLLTDISNSTFEWEVTTSDDIT
GFPEGVQTTTTIPSMPLINTCNTSGVVTIQVTPFNDGCEGLPVEFQITVNPKPFIFDLNTTICSGDTFLIEPNNDCANGQIVPENTTYNWVVDSQGPDLNGESGSGTSISQSLNNISDSVQSVVYLVTPTSGDTGSCLGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLN
GLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPL
VASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIG
YTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAG
QAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVT
VTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTS
EGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANS
VDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALI
NDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAG
ESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIV
DNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVNPEPVVLSISDIEVCTGEIVDTIIFSTTNT
GGVISYNWEINTDIGLLPLSVVGDIPSFTSQNSSNVPLVATVDVTPTFTANGLSCQGLSESFQIVVNPRPLINDKIVEICSEDTFSIQPLNNIDGDVVPNGTEYSWQVLLPNTNITGATSGSGTIISQTLINTSNEAQVVVYEVTPISNAIGACEGSTFEIEVTVTPRPFLQNQNFETCSGDTFTFSPIN
NPPNTIIPIGTTYTWTFTNNPNITGETDGINEAQFEQLNLINSTNVDQSVVYNITATAASCSYSFVIELVVKPTPFIPYNSGLTDTRCSGDPFIIQPVNGVPDSSTIVPINTTYTWVVIPNPNISGWSDNSTPVGLISQELYNLTNVNQSIEYIVTPYNGTCVGQTFSVLIWIEPKPQIPNHIETLCDGD
SFVFAPVNGVFPDSKTIVPDLTLYSWSVNDLSGGLISGYSNGVDQPFIDTGILNNDSNSVQTLIYDVTPTYYVNSNPNMPQCIGDTFTLTVSVNPGVEANAVVTNISCSYSPLCGGSIELNPSGVGPYTFNWTYSGSEINAISDPTLQNQYNLCPGDYMVEITDVFGCTYEFSYVIVPPQPVTFSLVSLV
DLSCNNISPNCDGYIEVDLQGGTLPYSLIEWYTESIPNSGNFDTIVSTGSNQLANACEGNYILKVLDTNACEFTSEIYTVEQTSSPIVINDQISDYNGYGVSCSNTNDGFITVDVLGGSGSFTYTLNPGDIQDSDPSTPNLLEFENLAPGAYTLDIIDSNCPQSITLNYNLTVPTAILSSHVLVSSPISC
YGGTETYTITASGGVPPYTGTGNYTLTAGTHDIVITDANGCSTTESIAVLEPTALTVNAAVTSSILCHGDSGIVTITASGGTPPYTGVGNITVSAGNCNYTVTDANGCSYSDTITVNEPDELLYTIDSTTNPTCSPDWSYTNGSICITITGGTNPIPTGTGWTDYGNGSWCLENLSAGDYAIDVTDDNGC
PSNGATDITLTRPPAIDAYITSNINADCSINTITQTNYVFVTGGTPPYEFSWSGGEVCDPINPQCMETTVSGTYTVYIHDQESLANGCPPIEVDVVVDLPEIGDAAFSYTSPNSILCDVLSFEEPISFFNESTGDIVNLTWDFGDGSMDVVGEENPTHVYSEIGTYQVELTVEYPYGCTETYSEVIEITK
GYDIVIPNAFTPNGDGINDTIRPVSLCMVQVKMSIYDTWGSLLYVEVGENDIINGWDGTIKGNPAENGNYIIVVEATTYRGETIELNGPITLIK
MKKNFILIFLFLTSLIGNAQQQSEDCIVPASLTLTHFNTAKYVPGGHLAVFFEPEGVFELDNEFVLELSDASGSFSTATTLSMKSEFFIPVLNGIIPLDIAPGSNYKLRIRTTAPFASVETDAFEIISQTTPDVVPAEITFINNAFTDLDNFIKCVDLAHDNYFFGHKDKGLTEVTPSDGGGIKLGLLGG
NSSNTVVRMLINGLWQELSITTIGSQFTIPSGLPVGYYLMELDKTVNPNTPNEYHNISGFIFHFNTNITGIANTSSETICVDQNAGFEVPIASIQYNYPGSMYSIHYGDSDETDTSTFDFYTHARLVNCNTLNHVYNATTCSSIFQDENPGNDNFYFKLDFNLYSEGLFNSDSDQFECETYTKNGGGTTK
WINASLKPSADLIAPDIICEGLAIVATDNSTSGLYGFGPECSSDYNTYWEVAAPGDDWVSVDPNNSIFTGWIDDVARTLTIPGSITSNNPGFWGIRLIISNPLGCVQSDTDEQTVIVEPIPAPSFDYTYQDNLCAPVTIEFTNTSNTEDINPPSFGNPTYTWSVVPDLTTPASLNGFTLLDENPNDNIDA
ANQTDIDIRFTQPGTYIVTLQLENECGTIFFPQNITIIGDPTVSFNPNSLEVCQEAPANYTLDFSDSTIQPLYSDTPYTPTTFLWEVFEIDGVTPASNYTFVNSTDAGTNYPQINFTEFGEYLIRITVSGNCEGNGSDDFLFNLKQTPIITNTNLTQEICSGDDTDEILLLTDISNSTFEWEVTTSDDIT
GFPEGVQTTTTIPSMPLINTCNTSGVVTIQVTPFNDGCEGLPVEFQITVNPKPFIFDLNTTICSGDTFLIEPNNDCANGQIVPENTTYNWVVDSQGPDLNGESGSGTSISQSLNNISDSVQSVVYLVTPTSGDTGSCLGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLN
GLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPL
VASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIG
YTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAG
QAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVT
VTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTS
EGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANS
VDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALI
NDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIVDNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAG
ESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVDPEPVVADQTLTVCSDVAIGYTIEGDVDTPVVATYNLTNVESNGLVGAGTNQGIENGLSSDALINDVWTNDTPSPVDVVYTLEPIGDNGCGGDEFTVTVTVDPEPVVADQAVTVCNDVAIGYTIQGDVDTPLVASYNLISIV
DNGLVGSTSNQGVGNTLPSDALTGDVWTNVTANSVDVLYTLVPIGDNGCEGDEFTLTVTVNPEPVVAGQAVTVCSDQALGIDFGSSSSVAATSYNITALTLNGLTVSSGGAGESSGLSASDLSDDAFTNTTPAAVDVVYTVVPVTSEGCEGEEFTLTVTVNPEPVVLSISDIEVCTGEIVDTIIFSTTNT
GGVISYNWEINTDIGLLPLSVVGDIPSFTSQNSSNVPLVATVDVTPTFTANGLSCQGLSESFQIVVNPRPLINDKIVEICSEDTFSIQPLNNIDGDVVPNGTEYSWQVLLPNTNITGATSGSGTIISQTLINTSNEAQVVVYEVTPISNAIGACEGSTFEIEVTVTPRPFLQNQNFETCSGDTFTFSPIN
NPPNTIIPIGTTYTWTFTNNPNITGETDGINEAQFEQLNLINSTNVDQSVVYNITATAASCSYSFVIELVVKPTPFIPYNSGLTDTRCSGDPFIIQPVNGVPDSSTIVPINTTYTWVVIPNPNISGWSDNSTPVGLISQELYNLTNVNQSIEYIVTPYNGTCVGQTFSVLIWIEPKPQIPNHIETLCDGD
SFVFAPVNGVFPDSKTIVPDLTLYSWSVNDLSGGLISGYSNGVDQPFIDTGILNNDSNSVQTLIYDVTPTYYVNSNPNMPQCIGDTFTLTVSVNPGVEANAVVTNISCSYSPLCGGSIELNPSGVGPYTFNWTYSGSEINAISDPTLQNQYNLCPGDYMVEITDVFGCTYEFSYVIVPPQPVTFSLVSLV
DLSCNNISPNCDGYIEVDLQGGTLPYSLIEWYTESIPNSGNFDTIVSTGSNQLANACEGNYILKVLDTNACEFTSEIYTVEQTSSPIVINDQISDYNGYGVSCSNTNDGFITVDVLGGSGSFTYTLNPGDIQDSDPSTPNLLEFENLAPGAYTLDIIDSNCPQSITLNYNLTVPTAILSSHVLVSSPISC
YGGTETYTITASGGVPPYTGTGNYTLTAGTHDIVITDANGCSTTESIAVLEPTALTVNAAVTSSILCHGDSGIVTITASGGTPPYTGVGNITVSAGNCNYTVTDANGCSYSDTITVNEPDELLYTIDSTTNPTCSPDWSYTNGSICITITGGTNPIPTGTGWTDYGNGSWCLENLSAGDYAIDVTDDNGC
PSNGATDITLTRPPAIDAYITSNINADCSINTITQTNYVFVTGGTPPYEFSWSGGEVCDPINPQCMETTVSGTYTVYIHDQESLANGCPPIEVDVVVDLPEIGDAAFSYTSPNSILCDVLSFEEPISFFNESTGDIVNLTWDFGDGSMDVVGEENPTHVYSEIGTYQVELTVEYPYGCTETYSEVIEITK
GYDIVIPNAFTPNGDGINDTIRPVSLCMVQVKMSIYDTWGSLLYVEVGENDIINGWDGTIKGNPAENGNYIIVVEATTYRGETIELNGPITLIK
Molecular weight: 427.39 kDa
Isoelectric point according different methods:
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5VWI7|A0A0C5VWI7_9FLAO 50S ribosomal protein L34
MSKRTFQPSKRKRRNKHGFRERMASANGRKVLARRRAKGRKKLSVSSESRHKK
MSKRTFQPSKRKRRNKHGFRERMASANGRKVLARRRAKGRKKLSVSSESRHKK
Molecular weight: 6.31 kDa
Isoelectric point according different methods:
Isoelectric point according different methods:
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
962,384 |
49 |
4,610 |
352.8 |
39.7 kDa |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
6.4 |
0.77 |
5.58 |
6.27 |
5.26 |
6.32 |
1.75 |
8.13 |
7.89 |
9.23 |
2.04 |
6.72 |
3.27 |
3.38 |
3.16 |
6.46 |
6.03 |
6.21 |
1.05 |
4.06 |
For dipeptide frequency statistics click here
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI: proteome isoelectric point database. Nucleic Acids Res. 2016. doi: 10.1093/nar/gkw978 |
Contact: Lukasz P. Kozlowski |

