Microbacterium sp. Root61
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinobacteria; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
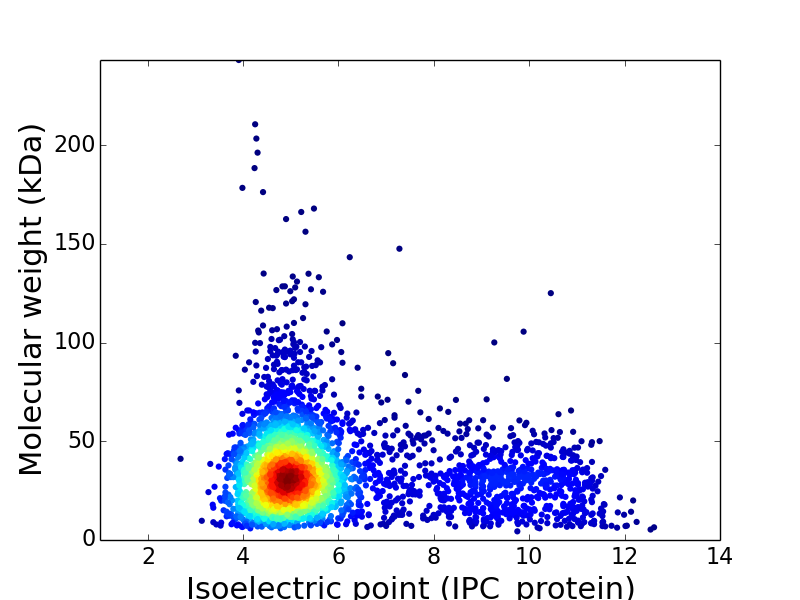
Virtual 2D-PAGE plot for 3,629 proteins (isoelectric point calculated using IPC_method)
Get csv file with sequences according given criteria:
* You can choose from 18 different methods for calculating isoelectric point
Protein with the lowest isoelectric point:
>tr|A0A0T2L1E1|A0A0T2L1E1_9MICO Uncharacterized protein
MAVNDNLNVLGIQTNLDTDLLSNNDVLSGNTDNSDNSDNSDNSDNSDNDFLSGNTDNSDNSDNSDNSDNSDNSDNSDNSDNSDNSDNSDNSVDTDVDVDVEDSFNEDDSYQDSINDSFQVNDSFQDNSIHDSANDNSVNAGIRQYNSGFGDINFIDDSEGGGGGMVGLMGAGSGGDFDFDVDARSTNNDQ
SVNQNITTNSGSGGAALAVGGDADGGFGGVLSGAGGGDSGNGGAGGSGTGAAGGAGGAGGLGGLGSGGAGGDGGLGGAGTGAAGGGSGGAGDAGSLGVGGGQGGSAGNANANGGDANGGNVSQFFNQDSVIVSGDGSYGAGGNVDVNNINTNIDVGDISIGNTWNTDSFNDDLDFSDNWSINDSFQDNSD
NSINDSFQDNSDDHGVDVDVDVDNSNVGSPFGDANEIDF
MAVNDNLNVLGIQTNLDTDLLSNNDVLSGNTDNSDNSDNSDNSDNSDNDFLSGNTDNSDNSDNSDNSDNSDNSDNSDNSDNSDNSDNSDNSVDTDVDVDVEDSFNEDDSYQDSINDSFQVNDSFQDNSIHDSANDNSVNAGIRQYNSGFGDINFIDDSEGGGGGMVGLMGAGSGGDFDFDVDARSTNNDQ
SVNQNITTNSGSGGAALAVGGDADGGFGGVLSGAGGGDSGNGGAGGSGTGAAGGAGGAGGLGGLGSGGAGGDGGLGGAGTGAAGGGSGGAGDAGSLGVGGGQGGSAGNANANGGDANGGNVSQFFNQDSVIVSGDGSYGAGGNVDVNNINTNIDVGDISIGNTWNTDSFNDDLDFSDNWSINDSFQDNSD
NSINDSFQDNSDDHGVDVDVDVDNSNVGSPFGDANEIDF
Molecular weight: 41.12 kDa
Isoelectric point according different methods:
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0T2KYN2|A0A0T2KYN2_9MICO Uncharacterized protein
MLGRRVGRVGVIGAPVAKAAVVGAAITPGPAPIAKAAVVGAAVTPGRSPVAKAAVVGAVATPRFRRI
MLGRRVGRVGVIGAPVAKAAVVGAAITPGPAPIAKAAVVGAAVTPGRSPVAKAAVVGAVATPRFRRI
Molecular weight: 6.42 kDa
Isoelectric point according different methods:
Isoelectric point according different methods:
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1,203,465 |
38 |
2,358 |
333.2 |
35.6 kDa |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
13.51 |
0.51 |
6.22 |
5.53 |
3.22 |
8.94 |
1.95 |
4.89 |
1.85 |
10.08 |
1.92 |
1.94 |
2.74 |
5.41 |
6.98 |
5.57 |
6.19 |
9.0 |
1.55 |
1.99 |
For dipeptide frequency statistics click here
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI: proteome isoelectric point database. Nucleic Acids Res. 2016. doi: 10.1093/nar/gkw978 |
Contact: Lukasz P. Kozlowski |

