Chitinophaga pinensis (strain ATCC 43595 / DSM 2588 / NCIB 11800 / UQM 2034)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Chitinophaga; Chitinophaga pinensis
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins
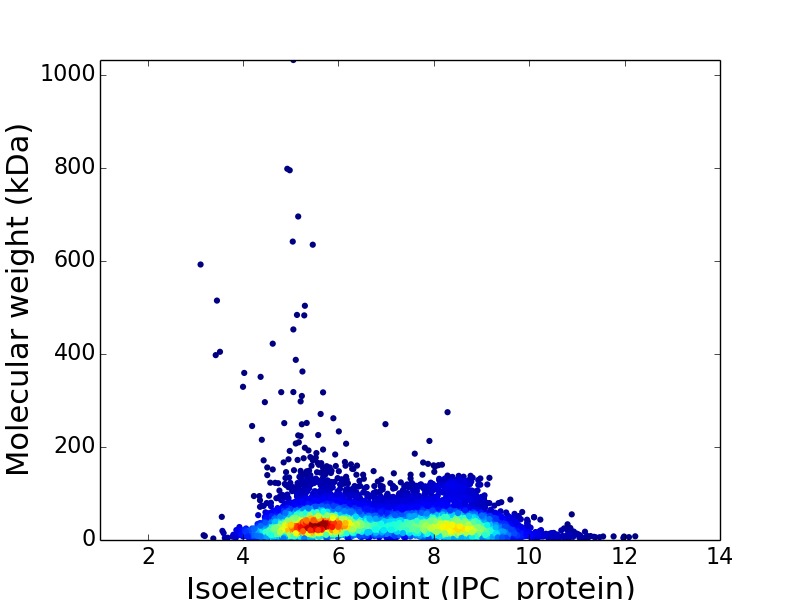
Virtual 2D-PAGE plot for 7,179 proteins (isoelectric point calculated using IPC_method)
Get csv file with sequences according given criteria:
* You can choose from 18 different methods for calculating isoelectric point
Protein with the lowest isoelectric point:
>tr|C7PA81|C7PA81_CHIPD Outer membrane adhesin like protein
MYQRQGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSAVVRFTVNAVNDAPVAVDDNVTVTEDVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSAVVRFTVNAVNDAPVAVDDNVTVTE
DVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGFPSLCDSAVVRFTVNAVNDAPVAVDDNVTVAEDVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSTVVRFTVNAVNDAPVAVDDNVTVTE
DVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSAVVRFTVNAVNDAPVAVDDNVTVTEDVPATGNVLTNDTDVEDNTLTASLVTAPVNGTIVLNADGSFTYAPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLITAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTVNPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALTASLVTAPVNGTVVLNAEGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGNSLSASLVTAPVNGTVALNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDVPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTINPVNDAPIAVDDQLTVVEDQTGSGNVLTNDSDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPVAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGNFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTINPVNDAPIAVDDQLTVVEDQTGSGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DIPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DIPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLIYQVCDNGTPSLCDTAVVRFTINPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITLTAANDAPLAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNTNYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDNDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTVNPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALTASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAIDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVLRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDNDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLIYQVCDNGTPSLCDTTVVRFTINPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFIYTPNTNYNGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPVAVDDQLTVIEDQIGTGNVLTNDTDPEGNSLTASLVNAPVNGTVVLNADGNFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVARFTINPVNDAPIAVDDQLTVVE
DQTGSGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVLRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGWDSLVYQVCDNGTPSLCDTAILRITVTAANDVPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLKADGSFTYTPNANYSGSDSLVYQVCDNGTPSLCDTAVVRFTINPVNDAPIAVDDQLTVVEDQTGSGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAADDQLTVVE
DQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGSDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAVDDQLTVIEDQIGIGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGNFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVARFTINPVNDVPIAVDDQLTVVE
DQTGSGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVLRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGWDSLVYQVCDNGTPSLCDTAILRITVTAANDVPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAADDQLTVVEDQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYGGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTAILVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPIAADDQLTVVEDQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLKADGSFTYTPNANYSGSDSLVYQVCDNGTPSLCDTAILRITVTPANDAPIAADDQLTVVE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPIAVDDQLTVIEDQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAVDDQLTVIE
DQIGTGNVLNNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSIIYRVCDNGTPSLCDTAILRITVTPVNDAPIAVDDQLTVVEDQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLNSDGSFTYIPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPVAADDQLTVVE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSMVYQVCDNGTPSLCDTAILRITVTPANDKPVAVDDVITVIEDQIGTGNVLTNDSDPEGDALSASVVKGPINGTIALNTDGSFTYTPNTNYSGLDSIIYRVCDNGTPSLCDTAVLRITVTPANDAPVAVDDQLTVVE
DQIGTGNVLTNDTDPEGDVLSASVVAGPVNGTIVLNADGSFTYTPNANYNGLDSVIYRVCDNGTPSLCDTAILRITVTPANDKPVAVDDAMTVVEDQTGTGNVLTNDTDPDGDALSASVVTGPVNGTIVLNADGSFTYTPNSNFNGLDSLIYRICDDGTPSLCDTAILRITVTSVNDAPTAVDDPLTVVE
DQTGTGNVLTNDSDPEGDALSASVVTGPVNGTIVLNADGSFTYTPNANYNGLDSVIYRVCDNGTPSLCDTAILRITVTPVNDKPIAVDDAVELMEDETATGNVMTNDSDPDGDPLTASLITAPVNGTFVLNADGSFTYTPNPNFNGADSMVYKICDNGTPALCDTAVVRFNILAVNDAPVAVNDIITVTE
DQPQNGNVLTNDSDPENDALSASVVAGPAHGTIVLNADGSFTYTPNANYNGMDTVTYRVCDNGTPSLCDTALIIITIVPGYDAPVAEADYLTVVEDTPTDGNALANDYDPDGDKLTATLVTAPVNGTVVFNADGTFTYTPNPDYEGPDSMVYRICDTDNLCDTAIIRITVIPGNEAPIATDDAVTINEDE
VATGNVLTNDTDPEGDALTASVITGPVNGTIVLNADGRFTYTPNTNYNGADSVIYQVCDNGSPSRCDTAVLRITIIPQNDPPVANIEIIRVEINTPIAGSVKPVISDPDGDPLTVSLIRATTHGVITLNADGSFEYSPNTDFSGTDSLIYKVCDNGTPSLCDTGIGIFNVSGPNDRPVIGIAKAAAEPVL
ELNGNYTVTYTFVVTNLGNEILNSVQVEDNLLNTFPSPHTFTISGDVVTTGSLRPNNQYNGSTISTLLGDNSTLAIGASDTIRFSVNVNTHKQFGTFNNSATATATSAGTSTEVTDVSTNGLNADPDGNADPSENEATPITLNPTKIRIPQGFSPNGDGKNDRFVIGNAGNDRINLEVYNRWGNVVYKNA
NYNNEWDGRCNYGLHFGENIPDGTYYIIVIVNDSERFVNYITVIR
MYQRQGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSAVVRFTVNAVNDAPVAVDDNVTVTEDVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSAVVRFTVNAVNDAPVAVDDNVTVTE
DVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGFPSLCDSAVVRFTVNAVNDAPVAVDDNVTVAEDVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSTVVRFTVNAVNDAPVAVDDNVTVTE
DVPATGNVLTNDTDVEGNTLTASLVTAPVNGTIVLNADGSFTYTPNANYNGLDSLVYQVCDNGVPSLCDSAVVRFTVNAVNDAPVAVDDNVTVTEDVPATGNVLTNDTDVEDNTLTASLVTAPVNGTIVLNADGSFTYAPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLITAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTVNPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALTASLVTAPVNGTVVLNAEGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGNSLSASLVTAPVNGTVALNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDVPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTINPVNDAPIAVDDQLTVVEDQTGSGNVLTNDSDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPVAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGNFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTINPVNDAPIAVDDQLTVVEDQTGSGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DIPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DIPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLIYQVCDNGTPSLCDTAVVRFTINPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITLTAANDAPLAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNTNYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDNDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVVRFTVNPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALTASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAIDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVLRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDNDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLIYQVCDNGTPSLCDTTVVRFTINPVNDAPIAADDQLTVIEDQTGSGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFIYTPNTNYNGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPVAVDDQLTVIEDQIGTGNVLTNDTDPEGNSLTASLVNAPVNGTVVLNADGNFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVARFTINPVNDAPIAVDDQLTVVE
DQTGSGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVLRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGWDSLVYQVCDNGTPSLCDTAILRITVTAANDVPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLKADGSFTYTPNANYSGSDSLVYQVCDNGTPSLCDTAVVRFTINPVNDAPIAVDDQLTVVEDQTGSGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAADDQLTVVE
DQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGSDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAVDDQLTVIEDQIGIGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGNFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVARFTINPVNDVPIAVDDQLTVVE
DQTGSGNVLTNDTDPEGNSLSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNVTVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAVLRITVTAANDAPVAVDDNVTVTE
DVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPVAVDDNETVTEDVPATGNVLTNDTDPEGDALSASLVTAPVNGTVVLNADGSFTYTPNANYNGWDSLVYQVCDNGTPSLCDTAILRITVTAANDVPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAADDQLTVVEDQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYGGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAVDDQLTVIE
DQIGTGNVLTNDTDPEGNSLTAILVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPIAADDQLTVVEDQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLKADGSFTYTPNANYSGSDSLVYQVCDNGTPSLCDTAILRITVTPANDAPIAADDQLTVVE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPIAVDDQLTVIEDQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTAANDAPIAVDDQLTVIE
DQIGTGNVLNNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYSGLDSIIYRVCDNGTPSLCDTAILRITVTPVNDAPIAVDDQLTVVEDQIGTGNVLANDTDPEGNSLTASLVTAPVNGTVVLNSDGSFTYIPNANYSGLDSLVYQVCDNGTPSLCDTAILRITVTPANDAPVAADDQLTVVE
DQIGTGNVLTNDTDPEGNSLTASLVTAPVNGTVVLNADGSFTYTPNANYNGLDSMVYQVCDNGTPSLCDTAILRITVTPANDKPVAVDDVITVIEDQIGTGNVLTNDSDPEGDALSASVVKGPINGTIALNTDGSFTYTPNTNYSGLDSIIYRVCDNGTPSLCDTAVLRITVTPANDAPVAVDDQLTVVE
DQIGTGNVLTNDTDPEGDVLSASVVAGPVNGTIVLNADGSFTYTPNANYNGLDSVIYRVCDNGTPSLCDTAILRITVTPANDKPVAVDDAMTVVEDQTGTGNVLTNDTDPDGDALSASVVTGPVNGTIVLNADGSFTYTPNSNFNGLDSLIYRICDDGTPSLCDTAILRITVTSVNDAPTAVDDPLTVVE
DQTGTGNVLTNDSDPEGDALSASVVTGPVNGTIVLNADGSFTYTPNANYNGLDSVIYRVCDNGTPSLCDTAILRITVTPVNDKPIAVDDAVELMEDETATGNVMTNDSDPDGDPLTASLITAPVNGTFVLNADGSFTYTPNPNFNGADSMVYKICDNGTPALCDTAVVRFNILAVNDAPVAVNDIITVTE
DQPQNGNVLTNDSDPENDALSASVVAGPAHGTIVLNADGSFTYTPNANYNGMDTVTYRVCDNGTPSLCDTALIIITIVPGYDAPVAEADYLTVVEDTPTDGNALANDYDPDGDKLTATLVTAPVNGTVVFNADGTFTYTPNPDYEGPDSMVYRICDTDNLCDTAIIRITVIPGNEAPIATDDAVTINEDE
VATGNVLTNDTDPEGDALTASVITGPVNGTIVLNADGRFTYTPNTNYNGADSVIYQVCDNGSPSRCDTAVLRITIIPQNDPPVANIEIIRVEINTPIAGSVKPVISDPDGDPLTVSLIRATTHGVITLNADGSFEYSPNTDFSGTDSLIYKVCDNGTPSLCDTGIGIFNVSGPNDRPVIGIAKAAAEPVL
ELNGNYTVTYTFVVTNLGNEILNSVQVEDNLLNTFPSPHTFTISGDVVTTGSLRPNNQYNGSTISTLLGDNSTLAIGASDTIRFSVNVNTHKQFGTFNNSATATATSAGTSTEVTDVSTNGLNADPDGNADPSENEATPITLNPTKIRIPQGFSPNGDGKNDRFVIGNAGNDRINLEVYNRWGNVVYKNA
NYNNEWDGRCNYGLHFGENIPDGTYYIIVIVNDSERFVNYITVIR
Molecular weight: 592.11 kDa
Isoelectric point according different methods:
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7PD58|C7PD58_CHIPD Uncharacterized protein
MKSLIMLMIAGLSLPGGKVYAQHVIVRVGSPPVLLTPRPAVIVAPRPVIVRPAPVVIVRPRRVVCRRPVLLVR
MKSLIMLMIAGLSLPGGKVYAQHVIVRVGSPPVLLTPRPAVIVAPRPVIVRPAPVVIVRPRRVVCRRPVLLVR
Molecular weight: 7.95 kDa
Isoelectric point according different methods:
Isoelectric point according different methods:
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2,669,891 |
30 |
9,175 |
373.4 |
41.8 kDa |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
8.11 |
0.84 |
5.37 |
5.48 |
4.52 |
6.96 |
2.04 |
6.61 |
5.95 |
9.52 |
2.37 |
5.12 |
4.09 |
4.12 |
4.48 |
6.2 |
6.19 |
6.53 |
1.28 |
4.23 |
For dipeptide frequency statistics click here
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI: proteome isoelectric point database. Nucleic Acids Res. 2016. doi: 10.1093/nar/gkw978 |
Contact: Lukasz P. Kozlowski |

